Research
Inside of our cells, genetic information is encoded on meter-long chromosomes with hundreds-of-millions of nucleotides organized inside micron-scale cell nuclei.
Our group uses computational approaches to identify and answer fundamental questions in genome biology posed by this immense range of scales. These include:
- How do molecular interactions between proteins organize micron-long chromosomes?
- Which DNA sequences specify when & how this organization occurs?
- How does genome organization influence communication between DNA regulatory elements?
We believe the answers to these questions are key links for connecting molecular processes and cellular phenotypes.
Our group currently develops:
- polymer simulations of genome folding mechanisms
- neural network models of DNA sequences determining genome folding
- computational tools for large-scale genomic data analysis and visualization
We often develop approaches using publicly-available datasets and work closely with collaborators to experimentally test predictions generated by analyses and models. Projects in the group span from model building to tool development to data analysis.
Two projects are highlighted below, with links to key papers, review articles, and code. The first project asks which DNA sequences specify genome organization and makes use of convolutional neural networks. The second aims to understand mechanisms of 3D genome organization, and makes use of polymer simulations.
DNA sequence determinants of 3D genome folding
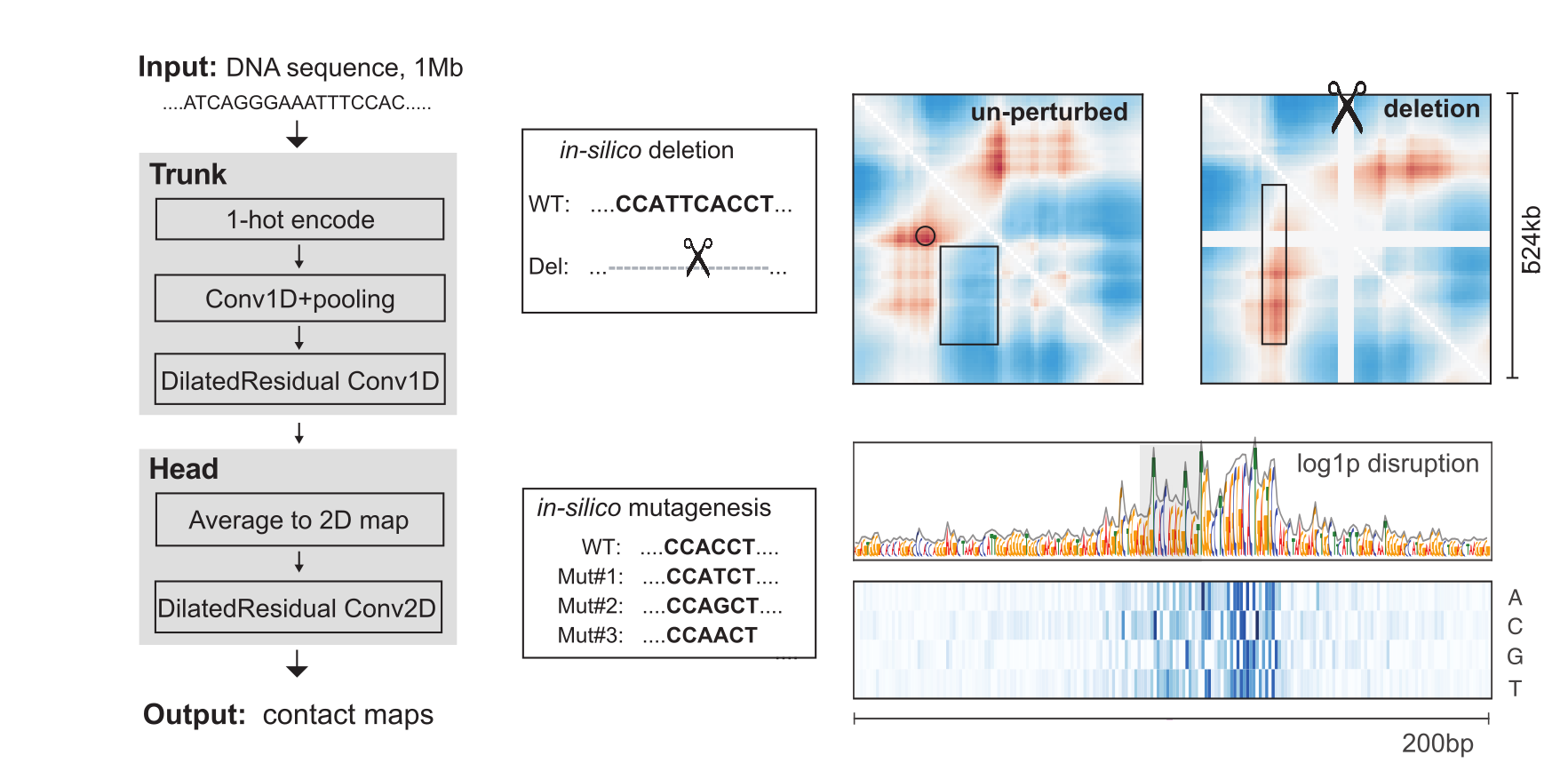
To connect the impact of individual DNA nucleotides with 3D genome folding, we leveraged recent progress in designing and training convolutional neural networks. We developed a convolutional neural network, Akita, that learns accurate representations of genome folding from DNA sequence (paper, github, poster & talk). The network has a ‘trunk’ which pools input sequences to 2048bp resolution and shares information across the sequence with dilated residual convolutions. This is followed by a ‘head’ which transforms from a latent space of 1D profiles to 2D maps of a megabase-by-megabase region. While computationally intensive to fit, trained models can be used to make hundreds of predictions per second, enabling in silico mutagenesis at scales beyond foreseeable experimental capabilities. We are excited about pursuing applications of this model as well as developing new network architectures and training strategies to make improved predictions. Applications we are excited about include those relating to: (i) evolution, (ii) de novo variant interpretation, and (iii) DNA sequence design in silico.
Loop extrusion as a mechanism of genome organization
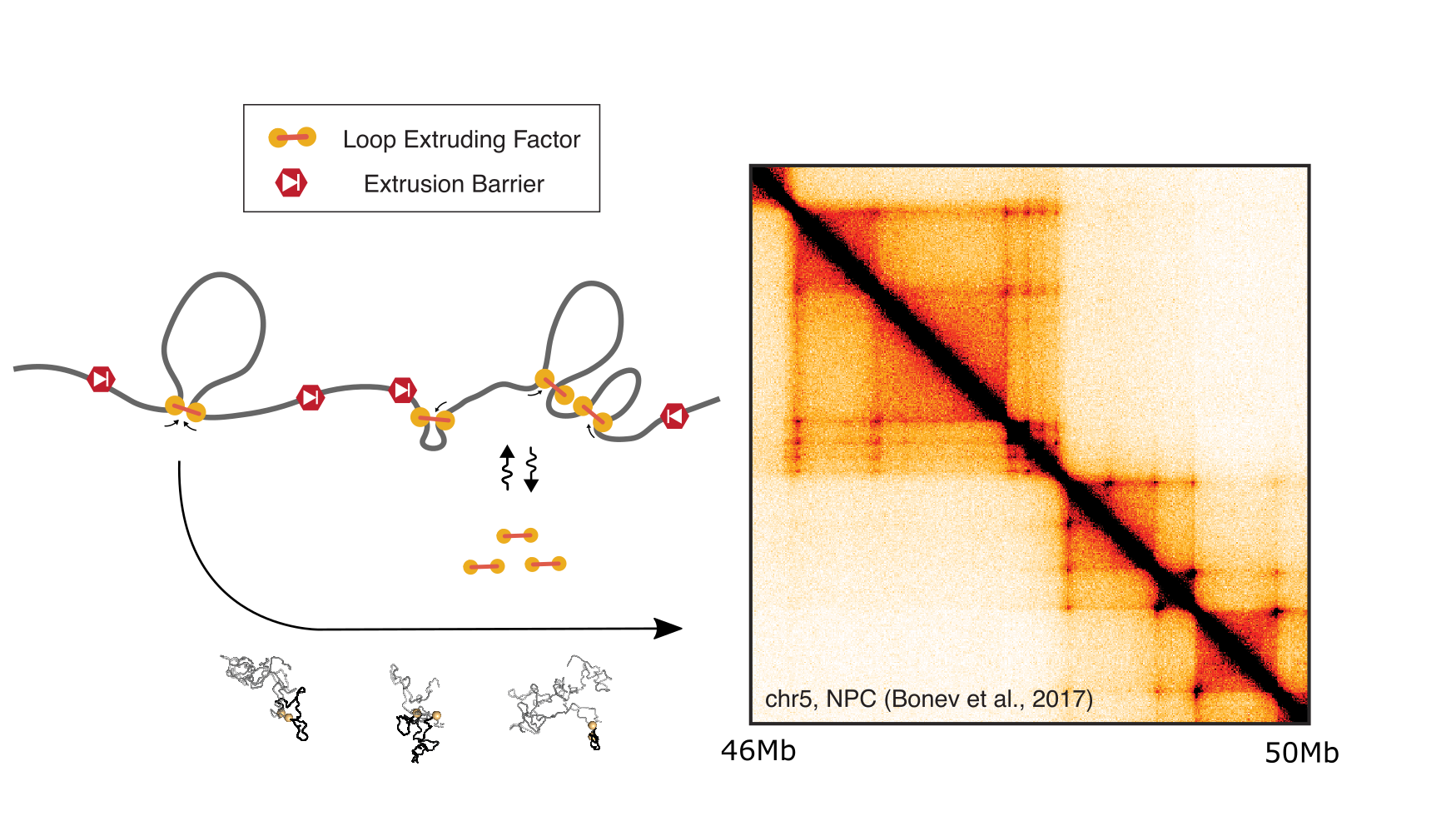
To understand how 3D folding patterns emerge in mammalian interphase Hi-C maps, we implemented and tested a range of possible mechanisms using polymer simulations. We found that the mechanism of loop extrusion limited by barriers could recapitulate many features of experimental data (paper, review article). This mechanism involves processive molecular machines that dynamically enlarge chromatin loops as they translocate hundreds of thousands of nucleotides along the chromatin fiber. Extruder translocation continues unless extruders become stalled at a barrier, or against another extruder, until they dissociate from the chromatin fiber. Spurred by these results, we collaborated with experimentalists to better understand the molecular basis for how barriers halt extrusion in interphase (paper), as well as how extrusion might operate to organize chromosomes in meiosis (paper). We are excited about developing new models to understand: (i) how loop extrusion can enable communication between enhancers and promoters, (ii) when and how regulators modulate loop extrusion dynamics, (iii) rules governing the encounters between extruders and barriers, (iv) the interplay between loop extrusion and other mechanisms of genome organization (e.g. heterochromatin phase separation in interphase, or synaptonemal complex assembly in meiosis).